Science Focus Area: Lawrence Livermore National Laboratory
- Principal Investigator: Yongqin Jiao1
- Laboratory Research Manager: Felice Lightstone1
- Co-Investigators: Dan Park1, Mimi Yung1, Jonathan Allen1, Jeff Kimbrel1, Adam Zemla1, Ali Navid1, Harris Wang2, William Bentley3, Gregory Payne3, Jeffrey Gralnick4, Jennifer Listgarten5
- Participating Institutions: 1Lawrence Livermore National Laboratory, 2Columbia University, 3University of Maryland, 4University of Minnesota, 5University of California–Berkeley
- Project Website: https://sc-programs.llnl.gov/biological-and-environmental-research-at-llnl/secure-biosystems-design
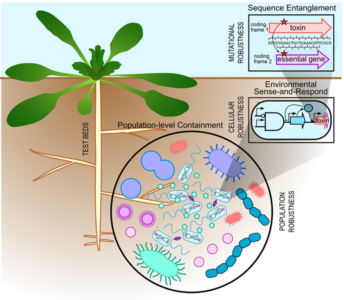
Microbial Secure Biosystems Design. The secure biosystems design Science Focus Area at Lawrence Livermore National Laboratory (LLNL) seeks to design multilayered containment strategies to safeguard the deployment of engineered microbes in the rhizosphere. [Courtesy LLNL]
Summary
Genetically engineered microorganisms (GEMs) play an important role in building and maintaining a sustainable bioeconomy. Without sufficiently robust biocontainment strategies, however, technology adoption and public trust will remain low. To reduce the risk of unintended ecological consequences from environmentally deployed GEMs, this Science Focus Area (SFA) is developing built-in security mechanisms that ensure GEM function where and when needed. Efforts are concentrated on establishing robust, generalizable biocontainment strategies in environmentally relevant soil microbes at the DNA sequence, cellular, and population levels to achieve containment without sacrificing microbial fitness. Leveraging Lawrence Livermore National Laboratory’s (LLNL) high-performance computing and high-throughput gene-editing capabilities, a specific focus of this SFA is on advancing a synthetic gene entanglement strategy for containment. In this strategy, two genes are encoded as overlapping sequences within the same DNA molecule, taking advantage of the fact that the same DNA sequence can be decoded in different reading frames. This DNA sequence overlap protects engineered functions against mutational inactivation and mitigates the potential transfer of engineered genes to naturally occurring microbes. Building on this layer of genetic stability, additional containment strategies that control cellular physiology and direct population coordination are incorporated to increase overall system robustness when confronted with environmental fluctuations. Leveraging LLNL’s rich experience in soil microbial ecology, this SFA evaluates the ecological benefits of these containment mechanisms in soil and rhizosphere environments. By increasing the genomic stability of GEMs and preventing the transfer of potentially invasive traits to native microbiomes, the ultimate objective is to achieve robust and secure niche-specific function of GEMs for safer and more effective environmental applications.
