Create open-access and integrated computational capabilities tailored to large-scale data science investigations for molecular, structural, genomic, and omics-enabled research on plants and microorganisms for a range of DOE mission goals.
Historically, systems biology computational infrastructure and software have been highly fragmented and lacked clear software development standards. Information streams are highly diverse, produced by many labs in a noncentralized manner, and are composed of vast quantities of data. To strengthen computational capabilities that broadly enable the research community, the Genomic Science Program (GSP) has been making a sustained investment in developing frameworks for open-access cyberinfrastructure for systems biology research through the Systems Biology Knowledgebase (KBase) project. Additionally, GSP is supporting efforts to make data objects FAIR (findable, accessible, interoperable, and reusable) through the National Microbiome Data Collaborative (NMDC) and Joint Genome Institute (JGI) data portals.
Looking into the future, BER will invest in and partner with other DOE Office of Science programs to provide access to enhanced mid-range and high-performance computing (HPC) facilities and cloud-based resources tailored to large-scale, integrative data science. As demonstrated during the COVID-19 pandemic, HPC-based analyses of integrated molecular, structural, genomic, and omics-enabled data can provide key insights into disease etiology and efficiently guide research toward potential treatment options out of myriad possibilities. This kind of powerful scientific inference, obtained from analyses of vast integrated datasets, needs to become a mainstay of GSP endeavors in Bioenergy Research, Biosystems Design, and Environmental Microbiome Research. GSP will build these capabilities into its portfolio, integrating them when practical and combining capabilities when necessary.
New Approaches for Large-Scale Biological Data Science
The availability of high-throughput, multiomics techniques is revolutionizing biological research, producing rich data layers of genomes, transcriptomes, proteomes, metabolomes, and other relevant data types. These data increasingly provide unparalleled insight into cellular processes, opening the door to significant advances in biological research (e.g., genome-scale engineering). However, the analysis of vast quantities of disparate data types is daunting, requiring new approaches for biological science.
DOE supports some of the most powerful computational systems and data transfer networks in the world, but these systems are not necessarily optimized for large-scale biological data science. Conducting next-generation biological research involves the use of big data, and the program’s ability to work efficiently and meaningfully with large datasets is increasingly important. Countries that develop the basic science infrastructure to work easily with large, complex datasets, particularly in the biosciences, will lead the world in bioenergy, biotechnology, synthetic biology, and microbiome science with enormous implications for human, economic, and environmental health. GSP seeks to develop online, open-access computational platforms for systems biology research.
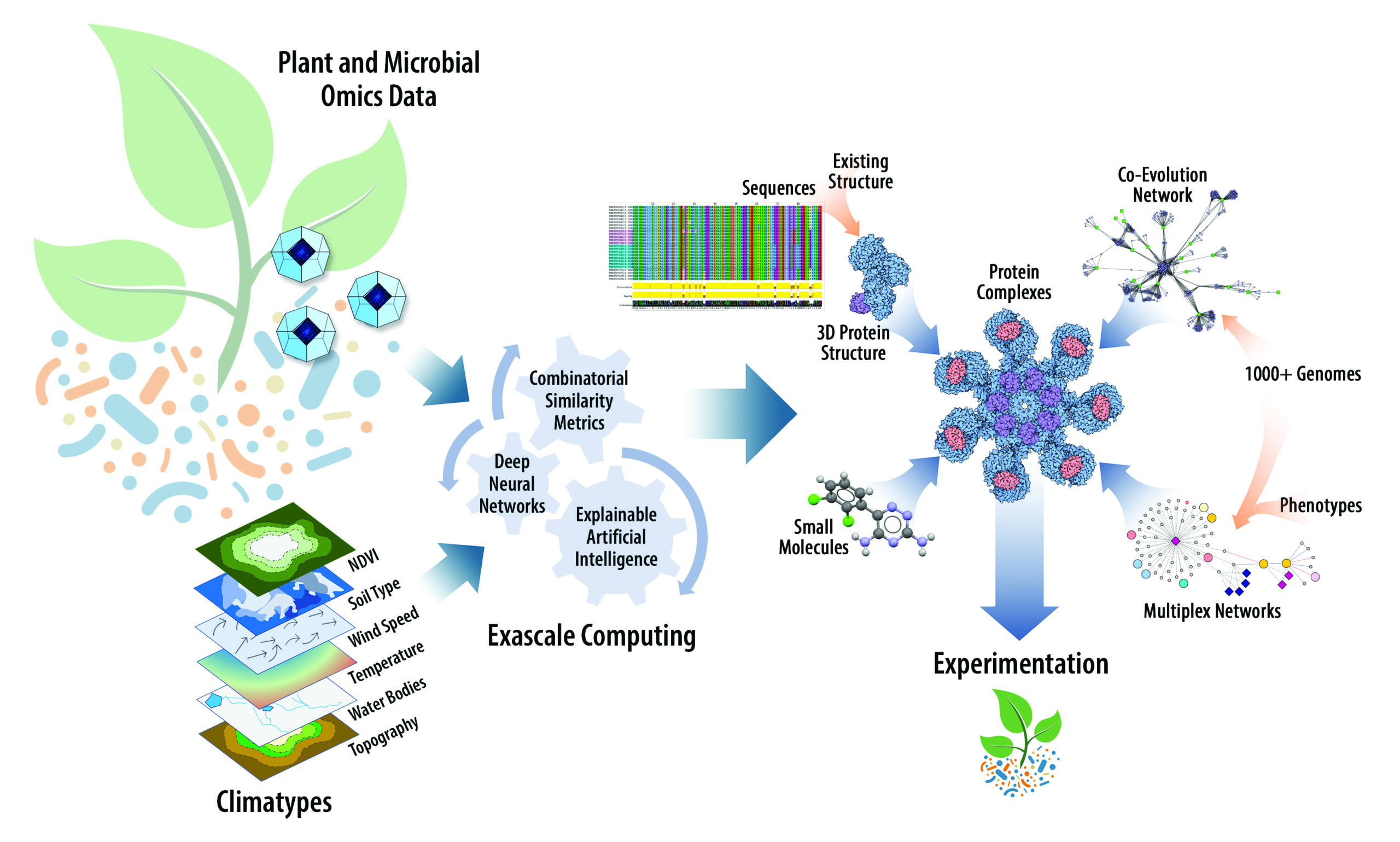
Integrated Systems Biology and High-Performance Computing Research Approach. Relationships within and across omics, phenomics, and environmental layers of data are captured with data-analytic and explainable artificial intelligence algorithms running on supercomputers and modeled as multiplex networks. Network-mining algorithms are then used to extract functional subnetworks of interest for mechanistic understanding of complex phenomena. Interactions among proteins and small molecules in these networks can be further modeled and explored in three dimensions. [Courtesy Oak Ridge National Laboratory, with portions reprinted with permission from Elsevier from Streich, J., et al. 2020. “Can Exascale Computing and Explainable Artificial Intelligence Applied to Plant Biology Deliver on the United Nations Sustainable Development Goals?” Current Opinion in Biotechnology 61, 217–25. DOI:10.1016/j.copbio.2020.01.010. © 2020.]
Other computational biology objectives are to:
- Assemble capabilities for processing large, complex, and heterogeneous systems biology data into open-access analysis platforms addressing DOE missions.
- Create the next-generation data systems and algorithms needed for large-scale systems biology data science that connects observations across scales and integrates molecular, structural, genomic, and other omics data with cellular and multicellular processes.
- Develop explainable artificial intelligence algorithms to identify relationships among different parts of genomes and build integrated biological models that capture higher-order complexity of the interactions among cellular components that lead to phenotypic differences.
- Generate advanced algorithms and data-handling techniques to process and integrate imaging and structural biology data with simulations and other biological measurements.
- Develop advanced simulation capabilities to model key processes at varying scales occurring within or among cells building toward whole-cell simulation.
- Assemble an integrated systems biology virtual laboratory to accelerate in silico ideation and collaboration within the research community.
Research in these areas will address significant lingering challenges, such as integrating datasets across varying spatial and temporal scales; extracting meaning from large, heterogeneous datasets; and linking three-dimensional structural protein data with network interaction models to predict protein interactions and small-molecule docking. Finding ways to efficiently solve current and future data challenges is a critical need for advancing BER science and will require close coordination with DOE’s Office of Advanced Scientific Computing Research and potentially other federal agencies. These computational efforts will leverage DOE’s HPC backbone, as future computing needs are anticipated to grow significantly.



