NLP for Synthetic Biology: Providing Generalizable Literature Mining Through KBase
Authors:
Shinjae Yoo1* (sjyoo@bnl.gov), Carlos X. Soto1, Gilchan Park1, Christopher Neely2, Vivek K. Mutalik2, and Paramvir S. Dehal2
Institutions:
1Brookhaven National Laboratory (BNL); and 2Lawrence Berkeley National Laboratory
Goals
The scientific literature contains many decades of research results which may inform the identification and subsequent engineering of microbial targets for novel applications, yet this knowledge remains largely inaccessible to current researchers due to the scale of the literature and the limitations of current manual information extraction practices using literature search and laborious manual curation processes used in String DB and AraNet. This project will produce a reusable proof-of-concept demonstration applying state-of-the-art natural language processing (NLP) techniques within the DOE Systems Biology Knowledgebase (KBase) framework to automatically extract organism traits from the literature for synthetic biology research. This work seeks to address important knowledge gaps in this field, while simultaneously providing a meaningful staging ground to expose new NLP tools to the KBase community and to gather feedback on their efficacy and use. The team will leverage NLP and data collection methods that have been previously developed and successfully applied in isolated settings to accomplish this effort, working with existing KBase tools and functionality, and producing outreach material to communicate and disseminate this work and its user-facing outcomes.
Abstract
Earth is facing some serious biological resource problems: a scarcity of renewable energy, lack of novel remedies for endemic infectious diseases, water pollution, shortages of arable soil and the resultant food crises, and the degradation of ecosystems to name a few of the most pressing. The project posits that the ability to domesticate and genetically engineer non-model microorganisms from relevant niches would help assess new potential solutions to many of these life-threatening global challenges. Though there have been technological innovations happening at rapid pace in addressing many of these challenges, the information for each potential new model organism is distributed throughout the literature and inaccessible to many practitioners, making unnecessarily difficult every new synthetic biology, bioenergy, and bioproduct project in a non-model organism. This lack of organized information not only limits machine-readable approaches, but also makes it difficult to assess the scope of work, identify knowledge gaps, and offer suggestions for investment to overcome technological barriers. For example, after decades of development in the field of synthetic biology, it is still challenging to identify suitable microbial targets for specific applications, conditions, and genetic tools necessary for the cultivation and engineering of non-model microorganisms. A generalized literature mining tool that keeps track of new technologies and genetic tools important for biotechnology practitioners would be invaluable. This tool will enable discovery of information gaps and opportunities that are buried within the vast literature. For example, the tool should be able to find information such as if the organism of choice is appropriate for domestication in the lab and which genetic tools exist for the organism with more ease than searching hundreds of primary literature sources, thereby saving time, effort, and money for the DOE-funded project. By building this prototype literature mining service into KBase, the team will be able to surface issues with KBase platform integration (including how the integration should be done), to identify the scoping and scaling needs, and to explore how to best address the needs of users.
Recently, improved techniques from the field of NLP have made it possible to analyze text at unprecedented scales (e.g., millions of documents), while extracting meaningful contextual information in ways not previously possible. These techniques are highly suitable to help address the above-mentioned knowledge gap. Here, the team will apply NLP to the biological literature to extract organism traits (see Figure 1) and deposit this mined knowledge in a useful form, supporting the creation of automated, curated centralized systems essential for growing and engineering of DOE-relevant microorganisms. While the BNL NLP techniques have extremely compelling applications, their value to BER researchers has been limited by access and dissemination of their results. KBase integration will bring the knowledge captured by the literature to a much wider audience.
Image
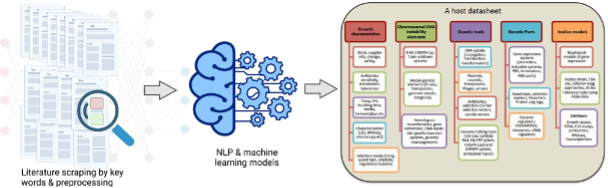
Figure 1. Overview of the project: The team proposes to use best natural language processing (NLP) and machine learning models to process and learn from literature data about growth characteristics, conditions, traits (such as antibiotic and stress tolerance, fitness traits, etc.), and available in silico models and genetic tools to engineer DOE-BER centric microorganisms.
Funding Information
This material is based upon work supported by the U.S. Department of Energy, Office of Science, Biological and Environmental Research (BER) Program, under Award Number DE-AC02-05CH11231 (LBL) and DE-SC-0012704 (BNL).
