Pilot Science Focus Area Project: Argonne National Laboratory
- Principal Investigator: Dionysios Antonopoulos1
- Co-Investigators: Gyorgy Babnigg1, Michael Fonstein1, Christopher Henry1, Michael Jewett2, Mark Mimee3, Arvind Ramanathan1, Yasuo Yoshikuni4
- Participating Institutions: 1Argonne National Laboratory, 2Northwestern University, 3University of Chicago, 4Lawrence Berkeley National Laboratory
Summary
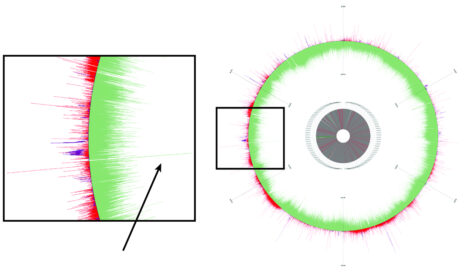
Secure Biodesign. Researchers are using CRISPR-enabled strategies to control specific populations of microorganisms and are leveraging machine-learning algorithms to better understand the precision of these mechanisms. Here, the activity of CRISPR guide RNA (gRNA) over time is overlaid on a depiction of the Escherichia coli MG1655 genome. Increased CRISPR gRNA activity is shown in green and decreased activity in red. The inset shows where CRISPR activity is particularly high in the middle of an intergenic region (arrow pointing to long green peak). [Courtesy ANL]
