AI-Informed Systems Biology: The Discovery of Cryptic Phenotypes and the Functional Networks that Control Them
Authors:
John Lagergren1,6* (lagergrenjh@ornl.gov), Mirko Pavicic1, Hari Chhetri1, Larry York1, Doug Hyatt2, David Kainer1, Erica Rutter3, Kevin Flores4, Jack Bailey-Bale5, Marie Klein5, Gail Taylor5, Jared Streich1, Daniel Jacobson1,6, and Gerald A. Tuskan1,6
Institutions:
1Oak Ridge National Laboratory; 2University of Tennessee–Knoxville; 3University of California–Merced; 4North Carolina State University; 5University of California–Davis; and 6Center for Bioenergy Innovation
URLs:
Goals
The Center for Bioenergy Innovation (CBI) vision is to accelerate domestication of bioenergy-relevant, nonmodel plants and microbes to enable high-impact innovations along the bioenergy and bioproduct supply chain while focusing on sustainable aviation fuels (SAF). CBI has four overarching innovation targets: (1) Develop sustainable, process-advantaged biomass feedstocks, (2) Refine consolidated bioprocessing with cotreatment to create fermentation intermediates, (3) Advance lignin valorization for biobased products and aviation fuel feedstocks, and (4) Improve catalytic upgrading for SAF blendstocks certification.
Abstract
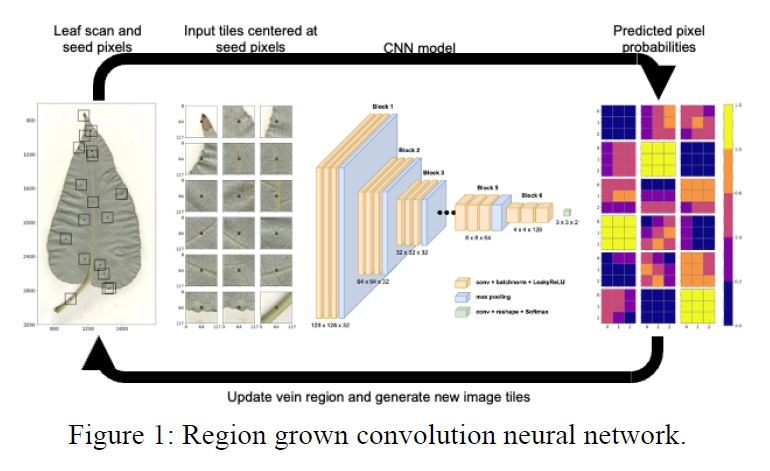
This project enables fast and accurate automated image-based plant phenotyping with minimal hand-annotated training data. Plant phenotyping is typically a time-consuming and expensive endeavor, requiring large groups of researchers to meticulously measure biologically relevant plant traits, and is one of the main bottlenecks in understanding plant adaptation and the genetic architecture underlying complex traits at population scale. Here the team addresses these challenges by leveraging few-shot learning with convolutional neural networks to segment the leaf body and visible venation of 2,906 P. trichocarpa leaf images obtained in the CBI common garden located at UC-Davis. In contrast to previous methods, the approach: (1) does not require experimental or image pre-processing, (2) uses the raw RGB images at full resolution, and (3) requires very few samples for training (e.g., just eight images for vein segmentation). Traits relating to leaf morphology and vein topology were extracted from the resulting segmentations using traditional image- processing tools and validated using real-world physical measurements.
To better understand the relationship among leaf phenotypes, a predictive phenomics network has been created from the leaf phenotypes with the use of iRF-LOOP, an explainable-AI-based network creation approach. Genome-wide association studies (GWAS) have been performed on each leaf phenotype and network-based functional partitioning has been performed across the GWAS results to determine the shared and distinct functional interactions responsible for governing leaf traits.
In this way, the current work provides the plant community with (1) methods for fast and accurate image-based feature extraction that require minimal training data, (2) a new population-scale phenotype data set, including 68 different leaf phenotypes, (3) a new SNP dataset for 1,419 genotypes called against v4.1 of the P. trichocarpa genome, and (4) a unique view of the functional relationships governing these leaf phenotypes. All few-shot learning code, data, and results are publicly available. This is one of the largest single releases of new plant genotype and phenotype data [www.osti.gov/dataexplorer/biblio/dataset/1846744].
Image
References
Cliff, A., et al. 2019. “A High-Performance Computing Implementation of Iterative Random Forest for the Creation of Predictive Expression Networks,” Genes 10(12), 996. DOI:10.3390/genes10120996.
Lagergren, J., et al. 2023. “Few-Shot Learning Enables Population-Scale Analysis of Leaf Traits in Populus trichocarpa,” arXiv preprint arXiv:2301.10351. DOI:10.48550/ARXIV.2301.10351.
Lagergren, J., et al. 2023. Supporting information for “Few-Shot Learning Enables Population-Scale Analysis of Leaf Traits in Populus trichocarpa,” Oak Ridge National Laboratory (ORNL), Oak Ridge, TN (United States). Oak Ridge Leadership Computing Facility (OLCF). DOI: 10.13139/ORNLNCCS/1908723.
Funding Information
Funding was provided by the Center for Bioenergy Innovation (CBI) led by Oak Ridge National Laboratory. CBI is funded as a U.S. Department of Energy Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the DOE Office of Science under FWP ERKP886. It also was supported by the Artificial Intelligence (AI) Initiative, an ORNL Laboratory Directed Research and Development program. This research used resources of the Oak Ridge Leadership Computing Facility, which is a DOE Office of Science User Facility. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the U.S. Department of Energy under contract no. DE-AC05-00OR22725.
