Develop a process-level understanding of microbiome function and be able to predict ecosystem impacts on the cycling of materials (carbon, nutrients, and contaminants) in the environment.
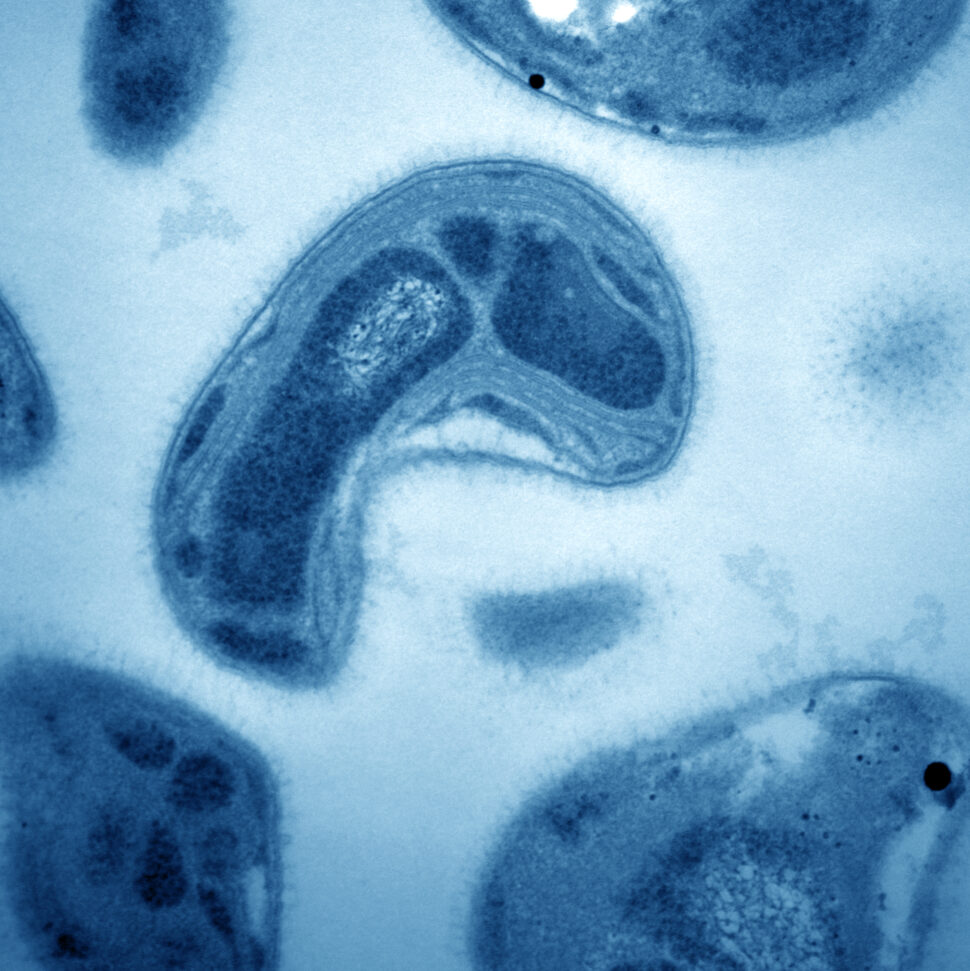
Methylosinus trichosporium. A transmission electron micrograph of Methylosinus trichosporium OB3b, a methanotroph that synthesizes a strong copper chelating compound – termed methanobactin – that prevents other microbes from collecting copper. Such competition for copper can cause soil greenhouse gas emissions to increase. [Courtesy Jeremy D. Semrau]
A key part of this understanding are microbial populations, which profoundly impact the world around us. They shape large-scale environmental processes (e.g., carbon, nitrogen, iron, phosphorus, and sulfur cycling), affect public health, and play an important role in industrial and biotechnology applications. Advances in the ability to measure, analyze, and manipulate microbiomes continue to provide exciting opportunities for scientific discovery. The Biological and Environmental Research (BER) Program has long been a vanguard of microbiome science by supporting transformational environmental research and investing in the development of cutting-edge technologies.
GSP will continue to build on this legacy by supporting omics-enabled basic research that tackles fundamental scientific and methodological challenges in microbiome science while maintaining an emphasis on the bioenergy, sustainability, and biogeochemical cycling elements of its research portfolio. Significantly, the breadth and complexity of biotic interactions that occur within and across microbiomes are not sufficiently captured by the current state of knowledge. Moreover, a persistent challenge in microbiome science is the ability to quantitatively model or temporally predict microbiome structure and behavior. GSP supports experimental and computational research that aims to understand microbial community interactions and biogeochemical cycling across broad time and space scales. In this context, the program supports studies that interrogate the vast, uncharacterized genomic diversity of environmental microbes, which represents an important bottleneck of discovery. Relevant efforts are closely tied to developing a better understanding of microbial impacts on sustainable bioenergy crop growth and yield. Finally, the program also seeks to leverage emerging opportunities in synthetic and computational biology by exploring the potential of synthetic microbiome science to advance predictive capabilities and to understand gene function across microbial taxa and ecosystems.

Understanding the Soil Microbiome. Kristen DeAngelis and Alex Bales sample soils from the Harvard Forest long-term warming experiment to understand how climate change affects microbial growth efficiency, soil carbon stocks, and feedbacks to the climate system. [Courtesy University of Massachusetts–Amherst]
- Support the development of environmental omics approaches (e.g., metagenomics, metatranscriptomics, metaproteomics, and community-scale metabolomics) and associated data integration tools to investigate in situ microbial community functional activities controlling key environmental processes.
- Facilitate realistic recapitulations of microbial ecosystems that move beyond the simple characterization of microbial diversity in the environment.
- Consider complex interkingdom biological dependencies among soil microbial community members (e.g., bacteria, fungi, archaea, viruses, and protists) and the spheres they inhabit (e.g., phycosphere and rhizosphere).
- Provide a framework for predictive modeling to understand microbial community function and dynamics by using omics data.
- Leverage omics and modeling approaches to understand the rate and magnitude of microbially mediated biogeochemical cycles (e.g., carbon, nitrogen, phosphorus, sulfur, and other elements) in the environment and to understand feedbacks with and responses to global change.
- Develop tools and techniques to manipulate microbiomes for beneficial purposes, such as enhanced sustainability, bioenergy crop productivity, and soil carbon storage.
- Facilitate novel and innovative approaches to functional gene characterization, including the use of data science tools (e.g., artificial intelligence and machine learning) and close integration with computational biology and infrastructure elements of GSP’s portfolio (e.g., KBase, JGI, and NMDC).
- Develop omics-enabled techniques for imaging microbial community structure and function in terrestrial environments, with an emphasis on technically challenging settings, such as soils, sediments, and key interfacial environments (e.g., decaying organic material, mineral aggregates, and plant roots), potentially by integrating these activities with BER’s biomolecular characterization and imaging science portfolio.
- Combine omics with in situ technologies that enable nondestructive, high-throughput sampling or measurements of microbial communities to understand process-level interactions.
- Seek a mechanistic understanding of cell-to-cell interactions, signaling, resource sharing, and communication at molecular, cellular, and community scales.
