Development of High-Throughput Light-Sheet Fluorescence Lifetime Microscopy for 3D Functional Imaging of Metabolic Pathways in Plants and Microorganisms
Authors:
Adam Bowman1* (abowman2@stanford.edu), Dara Dowlatshahi1,2, Franz Pfanner1, Soichi Wakatsuki1,2, and Mark Kasevich1
Institutions:
1Stanford University; and 2SLAC National Accelerator Laboratory
Goals
The goal is to realize a high-speed lifetime imaging platform for light-sheet microscopy of metabolic pathways and plant-microbe interactions using electro-optic fluorescence lifetime microscopy (EO-FLIM; Bowman and Kasevich 2021; Bowman et al. 2019). Wide-field optical modulators allow efficient lifetime capture combined with low noise readout on standard scientific cameras. This system will find broad applications in plant imaging and will provide lifetime contrast using both fluorescent labels and endogenous autofluorescence. Multi-dimensional imaging optics enable lifetime multiplexing and unmixing of autofluorescent signatures. These optics allow simultaneous acquisition of space, polarization, nanosecond time, and wavelength.
Abstract
Researchers have completed the design and construction of two custom microscopes for EO-FLIM imaging. The first microscope [Fig. 1(a, b)] allows multi-dimensional wide-field FLIM and will be expanded in the future for 2-photon lightsheet excitation. Its design includes two 40 MHz resonant Pockels cells and a compact multi-spectral unit (MSU) for simultaneous FLIM imaging in several spectral bands. The MSU is broadly applicable to any microscope, and in the system, it enables ongoing experiments with multi-label and autofluorescence unmixing. The microscope is equipped with a supercontinuum laser source for multi-band excitation and also a doubled Ti:Sapphire laser and pulse-picker for ultraviolet excitation. The second microscope (Fig. 1c) is a lightsheet platform for large field-of-view FLIM imaging. The resonant Pockels cell for this system is driven at 80 MHz and provides a 17 mm aperture for imaging. Imaging optics have been optimized and initial volume acquisitions are underway.
Critical to both systems is an efficient data analysis pipeline. Previous work has primarily focused on rapid single-frame lifetime estimation using a single camera exposure. By combining multiple exposures taken at different Pockels cell drive phases, it is also possible to extract multi-exponential information. Multi-phase EO-FLIM data is well suited to phasor analysis to study multi-exponential decays without fitting or histogram binning. Researchers are now implementing phasor analysis on EO-FLIM datasets to allow real-time display of lifetime data as it is acquired [Fig. 1(e, f)]. Phasor analysis will also enable multi-label unmixing and visualization of lifetime shifts from autofluorescent species upon binding to different substrates.
Several biological samples have been developed for FLIM imaging. A collection of engineered Pseudomonas putida cells, each with a different fluorescent protein of choice, have been generated covering a large spectral range to be used for unmixing FLIM signals from a population of bacterial cells containing different fluorescent proteins. The same strains are being used for phasor analysis of outer membrane vesicles. eGFP fused with tetraspanin Tet-8 serves as a marker of vesicles in Arabidopsis plants for live FLIM imaging of the root hair cells (Fig. 1d). Two carbon cycling enzymes, 4-hydroxybutyl-CoA dehydrogenase (4HBD) and enoyl-CoA reductase/carboxylase (ECR), as well as glucose-6-phosphate dehydrogenase (G6PD) have been expressed and purified with and without autofluorescent molecules, NADPH and/or FAD, for establishing FLIM and phasor signatures (Fig. 1e), which will be used for unmixing multidimensional live imaging of cells expressing these enzymes.
Researchers have also applied the EO-FLIM optics to kilohertz rate high-speed FLIM imaging of a FRET-based genetically encoded voltage sensor in Drosophila, enabling lifetime detection of action potentials in vivo. Lifetime readout significantly improves the signal-to-noise and stability of voltage recordings (Bowman et al. 2023). This work enables future directions for imaging dynamic signals throughout plants.
Image
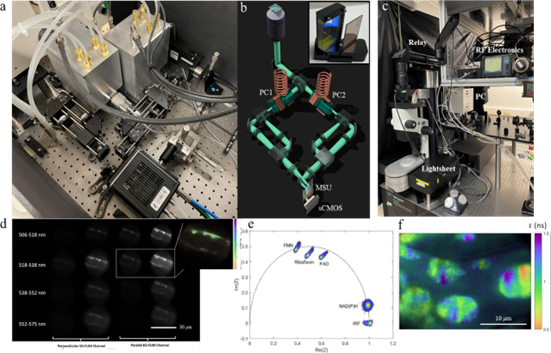
Figure 1: EO-FLIM optics and data (a, b) Multidimensional electro-optic fluorescence lifetime microscopy (EO-FLIM) optics using two resonant Pockels cells (b, inset). Multi-spectral unit (MSU) uses a stack of dichroic filters to create four spectrally separated bands on an sCMOS camera sensor as shown in (d). (c) EO-FLIM optics developed for lightsheet microscopy with wide field-of-view, including an 80 MHz resonant large-aperture Pockels cell (d) 4x4 array of images output from multidimensional optics encoding 4 spectral channels as rows and both lifetime and polarization information as columns. Here researchers are imaging vesicle transport in an Arabidopsis root hair. (inset) Lifetime image calculated from the ratio of one pair of output images in the array. (e) Phasor plot showing the signatures of various autofluorescence species excited with ultraviolet light (f) Lifetime image of structures in a plant leaf generated using phasor analysis.
References
Bowman, A. J., and M. A. Kasevich. 2021. “Resonant Electro-optic Imaging for Microscopy at Nanosecond Resolution.” ACS Nano 15(10), 16043–54. DOI: 10.1021/acsnano.1c0447.
Bowman, A. J. et al. 2019. “Electro-Optic Imaging Enables Efficient Wide-Field Fluorescence Lifetime Microscopy.” Nature Communications 10(1). DOI: 10.1038/s41467-019-12535-5.
Bowman, A. J., et al. 2023. “Wide-Field Fluorescence Lifetime Imaging of Neuron Spiking and Sub-Threshold Activity in vivo.” arXiv, 2211, 11229.
Funding Information
This research was supported by the DOE Office of Science, through the Biomolecular Characterization and Imaging Sciences Program, Office of Biological and Environmental Research (BER), grant no. DE-SC0021976.
