National Laboratory Project: Sandia National Laboratories
- Principal Investigator: Joe Schoeniger1
- Co-Investigators: Kelly William1, Catharine Mageeney1, Jesse Cahill1, Peter Otoupa1, Michael Jewett2, Farren Isaacs3, Jennifer Doudna4
- Participating Institutions: 1Sandia National Laboratories, 2Northwestern University, 3Yale University, 4University of California–Berkeley
Summary
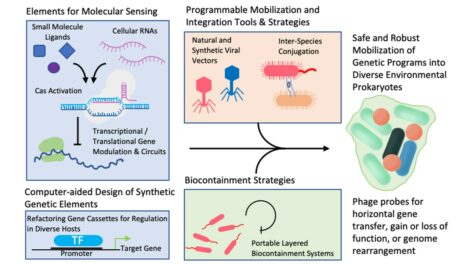
Microbial Community Composition. InCoGenTEC researchers are developing technologies to enable the study of changes in microbial community composition and genetics and the engineering of strong, multilayered biocontainment. Led by Sandia National Laboratories (SNL), the project’s technical themes include molecular tools for sensing and regulatory control, flexible genetic cassette design, bacteriophage vectors, and strategies for mobilizing and controlling genetic content. [Courtesy Northwestern University and SNL]
