02/08/2021
Unlocking a High-Quality Reference Genome for Switchgrass
Collaborative team including JGI and the four BRCs publishes complex genome analysis of switchgrass
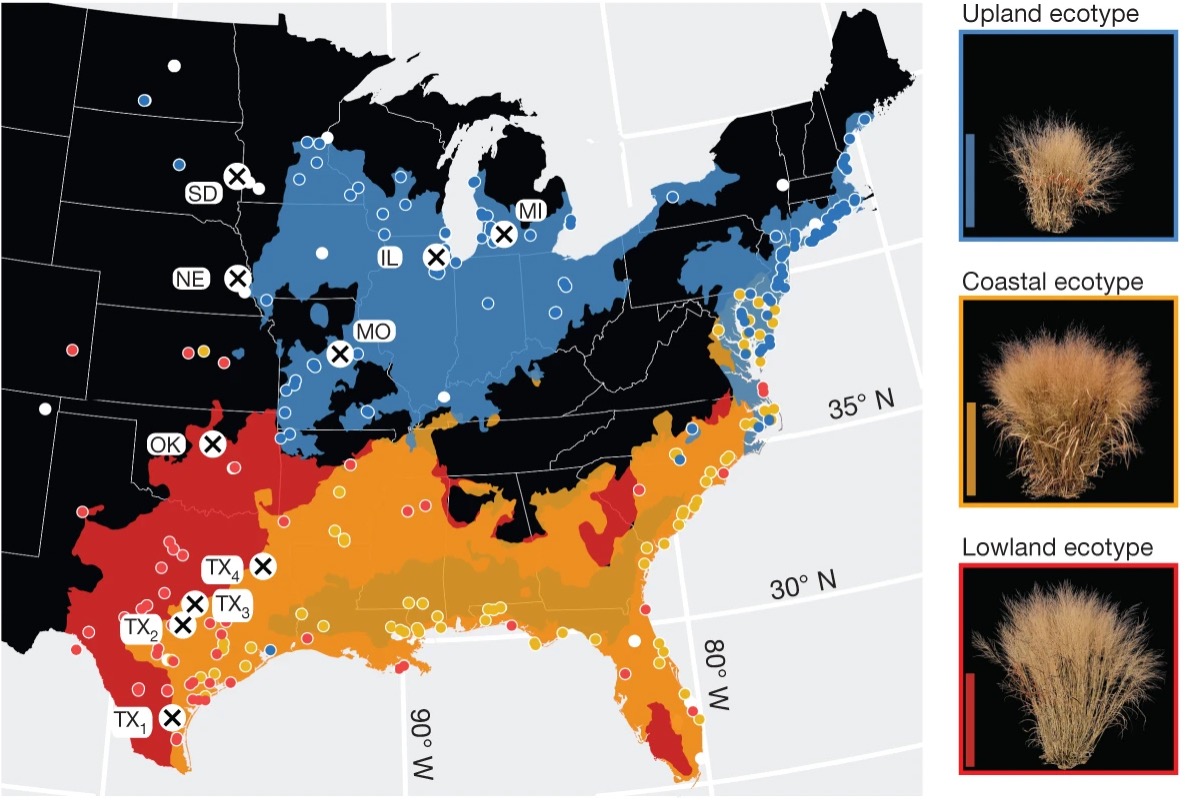
Geographic distribution of common gardens and plant collection locations, along with spatial distribution models of each ecotype. The ecotype color legend accompanies the representative images of each ecotype to the right of the map.
[Reprinted from Lovell, J.T. et al. 2021 under a Creative Commons license 4.0 International (CC BY 4.0).]
The Science
Switchgrass (Panicum virgatum) is a promising and versatile candidate for producing sustainable alternative fuels. However, outside their native habitat or cold hardiness zones, individual varieties of switchgrass tend not to be as productive. In the face of climate change, breeding strategies that can improve crop resilience and productivity across a range of environments are increasingly important, but these approaches require sufficient knowledge of the genes that underlie productivity and adaptation. By studying 732 genotypes grown at 10 experimental gardens in eight states spread across 1,100 miles, a large team of researchers have produced a high-quality reference sequence of the complex switchgrass genome.
The Impact
The reference genome will provide breeders with the necessary tools to increase switchgrass yield for the sustainable production of bioenergy. This research shows the power and value of the scientific collaborations made possible by the U.S. Department of Energy (DOE). This ambitious project, led in part by the Joint Genome Institute (JGI), a DOE Office of Science user facility located at Lawrence Berkeley National Laboratory, featured authors from university collaborators and all four DOE Bioenergy Research Centers (BRCs)—the Great Lakes Bioenergy Research Center (GLBRC), the Center for Bioenergy Innovation, the Center for Advanced Bioenergy and Bioproducts Innovation, and the Joint BioEnergy Institute.
Summary
Switchgrass is a perennial grass with a large and complex genome that has adapted to grow in a variety of soils, water conditions, and climates. The BRC program has developed switchgrass as a feedstock for plant-based fuels since 2007 and initiated work on sequencing the switchgrass genome more than a decade ago. This large and collaborative research team set up 10 experimental gardens in eight states spread across 1,100 miles, each containing a propagated panel of 732 switchgrass genotypes.
The genetic diversity across the set of plants allowed researchers to test which genes affect the plant’s adaptability to various environmental conditions. The combination of field data and genetic information has allowed the research team to associate climate adaptations with switchgrass biology, information that may help guide efforts to develop the crop as a versatile candidate biomass feedstock for producing sustainable alternative fuels. Building off this research, all four BRCs have expanded the network of common gardens and are exploring improvements to switchgrass through more targeted genome editing techniques to improve crop traits and customize the crop for additional end products.
This research was led by scientists at the University of Texas at Austin, the HudsonAlpha Institute for Biotechnology, and JGI. Also involved were researchers from the University of California–Berkeley, Rutgers University, U.S. Department of Agriculture’s (USDA) Agricultural Research Service, Arizona Genomics Institute, University of Georgia–Athens, Clemson University, Marshall University, Jawaharlal Nehru University, Noble Research Institute, University of Nebraska–Lincoln, South Dakota State University, University of Missouri, Argonne National Laboratory, USDA’s National Resources Conservation Service, Texas A&M University, University of California–Davis, Oklahoma State University, University of Oklahoma, and Washington State University.
This highlight is courtesy of, and originally published by, the Great Lakes Bioenergy Research Center.
Principal Investigator
David B. Lowry
Michigan State University
dlowry@msu.edu
BER Program Manager
Kent Peters
U.S. Department of Energy, Biological and Environmental Research (SC-33)
Biological Systems Science Division
kent.peters@science.doe.gov
Funding
This research was supported by DOE awards to T.E.J. (DESC0014156), D.B.L. (DESC0014156), K.M.D. (DE-SC0010743), the Great Lakes Bioenergy Research Center (DESC0018409 and DE-FC02-07ER64494), and the Center for Bioenergy Innovation (DE-AC05-000R22725). Funding was also provided by the National Science Foundation’s Genome Research Program to T.E.J. (awards IOS0922457 and IOS1444533) and J.T.L (IOS1402393). The DOE Office of Science supported the work by JGI (contract DE-AC02-05CH11231), the Joint BioEnergy Institute (contract DE-AC02-05CH11231), and Argonne National Laboratory (contract DE-AC02-06CH11357).
References
Lovell, J.T. et al. “Genomic Mechanisms of Climate Adaptation in Polyploid Bioenergy Switchgrass.” Nature 590, 438–444 (2021). [DOI:10.1038/s41586-020-03127-1]
