Populus Secondary Cell Walls: Atomistic Models and Natural Variability in Architecture Informed by Solid-State Nuclear Magnetic Resonance
Authors:
Bennett Addison1, Lintao Bu1, Vivek Bharadwaj1, Meagan Crowley1, Anne E. Harman-Ware1,2*(anne.ware@nrel.gov), Mike Crowley1, Peter Ciesielski1, Yannick J. Bomble1, and Gerald A. Tuskan2
Institutions:
1National Renewable Energy Laboratory; and 2Center for Bioenergy Innovation
URLs:
Goals
The Center for Bioenergy Innovation (CBI) vision is to accelerate domestication of bioenergy-relevant, nonmodel plants and microbes to enable high-impact innovations along the bioenergy and bioproduct supply chain while focusing on sustainable aviation fuels (SAF). CBI has four overarching innovation targets: (1) Develop sustainable, process-advantaged biomass feedstocks, (2) Refine consolidated bioprocessing with cotreatment to create fermentation intermediates, (3) Advance lignin valorization for bio-based products and aviation fuel feedstocks, and (4) Improve catalytic upgrading for SAF blendstocks certification.
Abstract
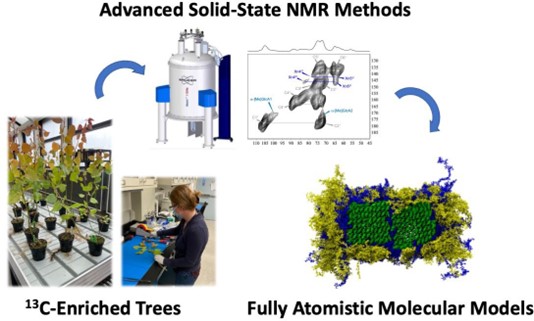
Interactions and higher-order architecture of the biopolymers present in plant secondary cell walls (SCW) are poorly understood. Solid-state Nuclear Magnetic Resonance (ssNMR) measurements were used to infer refined details regarding the arrangements of the three major biopolymers in Populus cell walls: lignin, cellulose, and hemicellulose. Atomistic, macromolecular models representing multiple plant secondary cell wall arrangements were constructed in silico. Quantitative observables from molecular dynamics (MD) simulations (e.g., polymer-polymer distances, conformational analyses, quantified surface contact) were compared to ssNMR data to identify the most plausible polymeric arrangements. Results further understanding of plant SCWs and enable the development of hypotheses regarding superstructural architecture and resulting physicochemical properties of biomass cell walls. Additionally, researchers evaluated the genotypic variability of biopolymer spatial proximities in Populus SCW from a selection of natural variants exhibiting a range of compositional phenotypes. Coupling understanding of variability in SCW architectural characteristics with predictive atomistic models corroborated by ssNMR measurements will enable optimized design and deconstruction of sustainable biomass feedstocks. A high-level summary of the process workflow is presented in the figure below.
Image
References
Addison, B., et al. 2020. “Selective One-Dimensional 13C–13C Spin-Diffusion Solid-State Nuclear Magnetic Resonance Methods to Probe Spatial Arrangements in Biopolymers Including Plant Cell Walls, Peptides, and Spider Silk,” J. Phys. Chem. B 124(44), 9870–9883. DOI:10.1021/acs.jpcb.0c07759.
Funding Information
Funding was provided by the Center for Bioenergy Innovation (CBI) led by Oak Ridge National Laboratory. CBI is funded as a U.S. Department of Energy Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the DOE Office of Science under FWP ERKP886. Oak Ridge National Laboratory is managed by UT- Battelle, LLC for the U.S. Department of Energy under contract no. DE-AC05-00OR22725. This research also was supported by the U.S. Department of Energy (DOE), Office of Energy Efficiency and Renewable Energy (EERE), Bioenergy Technologies Office (BETO), under award no. DE- AC36-08GO28308 with the National Renewable Energy Laboratory. The molecular modeling contributions to this work were performed as part of the Feedstock Conversion Interface Consortium (FCIC) with funding graciously provided by the U.S. Department of Energy Bioenergy Technologies Office.
