Research Approach
Fundamental genomic science research includes single-investigator projects, multi-institutional collaborations, and research centers at universities and national laboratories across the country.
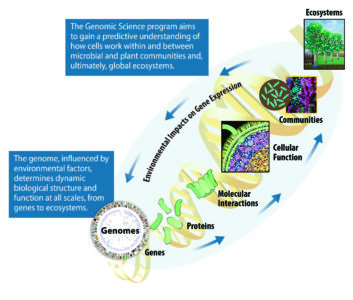
Multiscale Explorations. Achieving a predictive understanding of fundamental life processes requires investigations that span multiple levels, from the information encoded in individual plant and microbial genomes to the functioning of cells as communities in an ecosystem. Important to this challenge is understanding the complex interactions between each level of biological organization and the environment.
Scope and Progress
High-throughput genome sequencing of microbes, plants, and complex environmental assemblages of organisms has provided the vital blueprint necessary to understand the functional potential of organisms and interactive communities. By examining the translation of genetic codes into integrated networks of regulatory elements, catalytic proteins, and metabolic networks that define all living organisms, systems biology research sheds light on the fundamental principles that govern functional properties of organisms and how their processes respond to community interactions and environmental variables.
The Genomic Science program supports systems biology research aimed at identifying these foundational principles driving biological systems of plants, microbes, and multi-species communities relevant to DOE missions in energy and the environment.
Genomic Science program research is conducted at national laboratories and universities and includes single-investigator projects, multi-institutional collaborations, and fundamental research centers. The genome sequences of organisms studied in these projects are provided largely by the DOE Joint Genome Institute (JGI), an important user facility and a world leader in generating sequences of microbes, microbial communities, plants, and other organisms.
Genomic Science Approaches
Addressing extremely complex science questions that span all scales of biology, research supported by the Genomic Science program requires the collective expertise of scientists from many disciplines and the coordinated application of a wide range of technologies and experimental approaches, including genomics and metagenomics, analytical “omics,” molecular imaging and structural analysis, predictive modeling, and genome-scale engineering.
Genomics and Metagenomics. Sequencing and analyzing DNA from individual organisms (genomics) or microbial communities in environmental samples (metagenomics) form the foundation for systems biology research. The DOE Joint Genome Institute is an important scientific user facility that generates high-quality sequences and analysis techniques for diverse microbes, plants, and other organisms relevant to DOE energy and environmental missions.
Analytical Omics. Transcriptomics, proteomics, metabolomics, and other analyses—collectively described as “omics”—identify and measure the abundance and fluxes of key molecular species indicative of organism or community activity. Global analyses of important cellular components such as RNA transcripts, proteins, and metabolites inform scientists about organisms’ physiological status. This research, along with chemical and structural analytical technologies including stable isotope tracking and nano secondary ion mass spectrometry (NanoSIMS), also provides insights into gene function and indicates which genes are activated and translated into functional proteins as organisms and communities develop or respond to environmental cues. Methods that analyze DNA, RNA, proteins, and other molecules extracted directly from environmental communities enable discovery of new biological processes and provide novel insights into relationships between the composition of communities and the functional processes that they perform.
Molecular Imaging and Structural Analysis. Genomic Science program investigators are developing and using new methods for characterizing the chemical reaction surfaces, organization, and structural components in molecular complexes and tracking molecules to view cellular processes as they are occurring. Depending on the spatial scale, a variety of imaging technologies can be used to visualize the complex molecular choreography within biological systems. Some of these tools (e.g., synchrotrons, neutron sources, and electron microscopes) are available at DOE Office of Science user facilities that provide state-of-the-art spatial, temporal, and chemical measurement sensitivity.
Predictive Modeling. Computational models are used to capture, integrate, and represent current knowledge of biology at various scales. Researchers are using genome sequences and molecular, spatial, and temporal data to build models of signaling networks, gene regulatory circuits, and metabolic pathways that can be iteratively tested and validated to refine system understanding.
Genome-Scale Engineering. Genomes and systems-level understanding are uncovering the principles that govern system behavior, enabling genome-scale redesign of organisms. This research approach may involve building entirely new microbes from a set of standard parts—genes, proteins, and metabolic pathways—or radically redesigning existing biological systems to enable capabilities that the systems would not possess naturally.
