Science Focus Area: Los Alamos National Laboratory
- Principal Investigator and Laboratory Research Manager: Patrick Chain1
- Technical Co-Manager: Aaron Robinson1
- Co-Investigators: Julia Kelliher1, Reid Longley1, Buck Hanson1, Demosthenes (Dean) Morales1, La Verne Gallegos-Graves1, Leah Johnson1, Michal Babinski1, Kelli Feeser1, James (Jim) Brunner1, Justine Macalindong1, Pilar Junier2, Saskia Bindschedler2, Guillaume Cailleau2, Melissa Cravero2, Gregory Bonito3, Megan Korne3, Scott Baker4, Mary Lipton4
- Former Members: Geoffrey House, Debora Rodrigues, Jamey Young, Jason Corwin, Jim Werner
- Unfunded Collaborators: Donald Nativg5, Jennifer Rudgers5, Po-E (Paul) Li1, Sanna Sevanto1, Migun Shakya1, Yan Xu1, Ari Jumpponen6, Karolina Michalska7, Zou Finfrock7
- Participating Institutions: 1Los Alamos National Laboratory, 2University of Neuchâtel, 3Michigan State University, 4Pacific Northwest National Laboratory, 5University of New Mexico, 6Kansas State University, 7Argonne National Laboratory
- Overview Brochure: Download PDF
- Project Website: https://science-innovation.lanl.gov/science-programs/office-of-science-programs/biological-and-environmental-research/sfa-bacteria-fungal/
- KBase Collaboration: Integrating fungal computational sequence data analysis capabilities
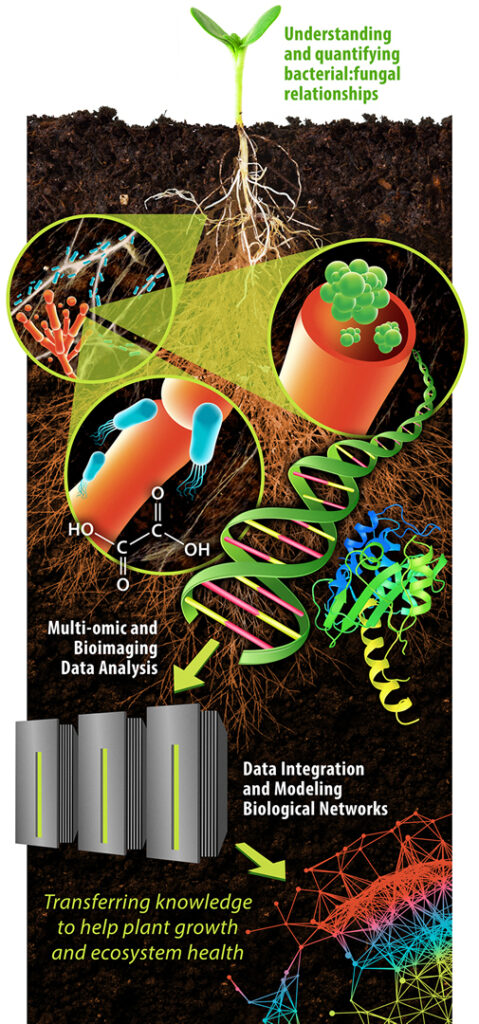
Understanding Bacterial-Fungal Interactions. SFA scientists are using a variety of approaches, including advanced microbiology, state-of-the-art omics, bioimaging techniques, and DOE computing capabilities, to understand the mechanisms driving the interactions and functions of soil bacteria and fungi. [Courtesy LANL]
Summary
Bacteria and fungi are the dominant players in soil microbiomes, with key roles in carbon flux, nutrient cycling, and plant productivity. This project aims to discover fundamental principles underlying bacterial:fungal interactions (BFI) to harness their biotechnological potential for potentially steering the function of soil ecosystems. The SFA’s 10-year vision is to predictively understand complex BFI, in the context of continued environmental change, to enable intentional steering of ecosystem functions for improved and sustainable bioenergy crop production and to enhance soil carbon sequestration and storage. Currently, the main goal is to better understand fundamental biological processes underlying highly specialized interactions between bacteria and fungi. This SFA uses novel bioinformatic algorithms with advanced scientific computing capabilities provided by the U.S. Department of Energy (DOE), coupled with targeted metagenomics to identify previously uncharacterized bacterial:fungal associations. Research will determine the range of BFI in the context of abiotic environmental changes such as shifts in moisture, nutrient availability, and temperature. The project also will characterize the underlying mechanisms of these interactions at the cellular and molecular scales using multiomics interrogation coupled with advanced in situ imaging techniques. These studies will provide the foundational knowledge for predictive models of soil community function that is required to fulfill the biotechnological potential of the soil microbiome.
