05/19/2022
Researchers Map Genetic Diversity in Switchgrass
Inter-ecotypic populations reveal quantitative trait loci underlying biomass yield
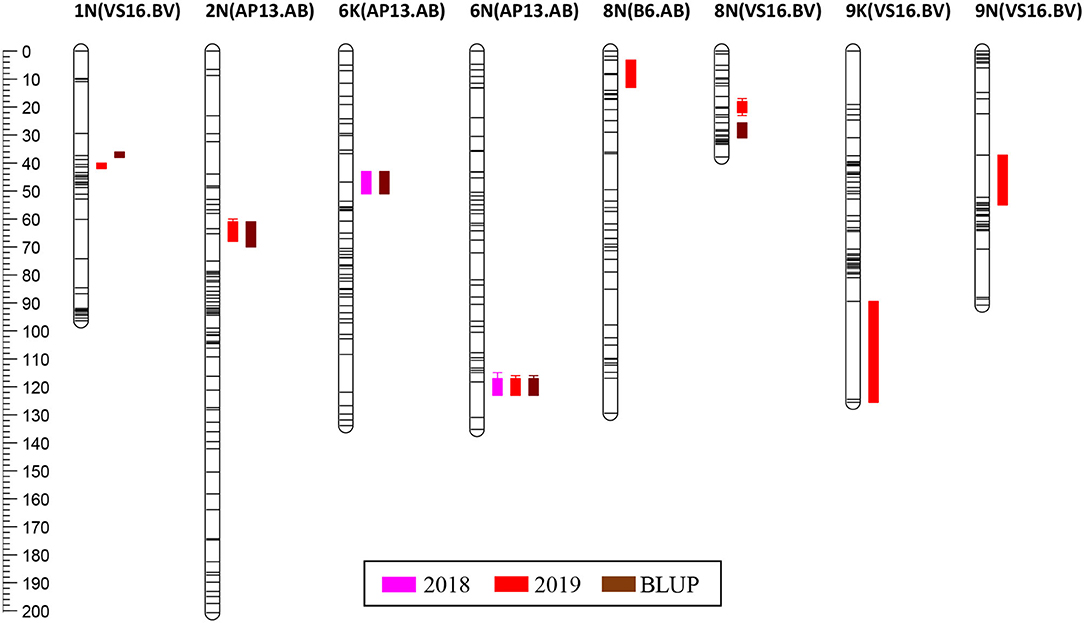
Positions of quantitative trait loci (QTLs) on a genetic map of switchgrass for harvest years 2018, 2019, and as estimated by the statistical method of best linear unbiased prediction (BLUP). Determining the location of the QTLs may potentially be used for marker-assisted improvement of biomass yield.
[Reprinted under a Creative Commons license (CC BY) from Razar et al. 2022. DOI: 10.3389/fpls.2022.739133]
The Science
The prevalence of genetic diversity in switchgrass germplasm can be exploited to capture favorable alleles that increase range of adaptation and biomass yield. Center for Bioenergy Innovation (CBI) researchers constructed genetic maps in two switchgrass F1 populations derived from different crosses between lowland, upland, and coastal switchgrass ecotypes. They analyzed polymorphisms and patterns of segregation distortion—a genetic deviation from expected Mendelian ratios which impacts interbreeding compatibility between ecotypes and drives evolution—and used linkage maps to locate quantitative trait loci (QTLs) for biomass yield. Wide crosses, such as between different ecotypes, can benefit switchgrass breeding efforts to improve growth and yield in unfavorable environmental conditions by selecting for desired genes and traits.
The Impact
Strong evidence for biomass yield QTLs indicate that breeding can improve switchgrass crops, especially across compatible inter-ecotypes. A challenge to achieving improved traits is the phenomenon of segregation distortion of alleles, which leads to unexpected trait inheritance outcomes. Transmission bias within individuals and loci needs to be considered as it may affect genetic gain when favorable alleles are distorted.
Summary
Two inter-specific ecotypic switchgrass F1 populations were generated: “AB” from a lowland (AP13) x coastal (B6) cross and “BV” from a coastal (B6) x upland (VS16) cross. The extent of genetic variance and patterns of segregation distortion were mapped for the two populations, with AB displaying more segregation distortion of alleles than BV, likely resulting from zygotic or postzygotic selection for increased heterozygosity. The results suggest lower genetic compatibility between lowland and coastal ecotypes than between coastal and upland ecotypes. Biomass yields were also analyzed across two years, and QTL mapping was performed to link genotypic and phenotypic information. Four QTLs for biomass yield were mapped in the AB population and six QTLs in the BV population, providing targets for switchgrass crop improvement.
Principal Investigator
Ali M. Missaoui
University of Georgia, Athens
cssamm@uga.edu
BER Program Manager
Shing Kwok
U.S. Department of Energy, Biological and Environmental Research (SC-33)
Biological Systems Science Division
shing.kwok@science.doe.gov
Funding
Partial funding was provided by the Center for Bioenergy Innovation, a DOE Bioenergy Research Center supported by the DOE Office of Science, Biological and Environmental Research program.
References
Razar, R., et al. 2022. “Genotyping-by-Sequencing and QTL Mapping of Biomass Yield in Two Switchgrass F1 Populations (Lowland x Coastal and Coastal x Upland),” Frontiers in Plant Science 13:739133. DOI: 10.3389/fpls.2022.739133.
